Illumina Sequencing Technology
868.95k views629 WordsCopy TextShare
Illumina
Watch the Updated Video: https://youtu.be/fCd6B5HRaZ8
This video provides an overview of the DNA se...
Video Transcript:
sample preparation begins with extracted and purified DNA the first step in next era sample preparation is tagged mentation during tag mminton transpose ohms simultaneously fragment and tag the input DNA with adapters once the adapters have been ligated reduce cycle amplification adds additional motifs such as the sequencing primer binding sites indices and regions that are complementary to the flow cell oligos clustering is a process wherein each fragment molecule is isothermally amplified the flow cell is a glass slide with lanes each lane is a channel coated with a lawn composed of two types of ala goes hybridization
is enabled by the first of the two types of ala goes on the surface this ala go is complementary to the adapter region on one of the fragment strands a polymerase creates a complement of the hybridized fragment the double-stranded molecule is denatured and the original template is washed away the strands are clonally amplified through bridge amplification in this process the Strand folds over and the adapter region hybridizes to the second type of ala go on the flow cell polymerases generate the complementary strand forming a double-stranded bridge this bridge is denatured resulting in two single-stranded copies
of the molecule that are tethered to the flow cell the process is then repeated over and over and occurs simultaneously for millions of clusters resulting in clonal amplification of all the fragments after bridge amplification the reverse strands are cleaved and washed off leaving only the forward strands the three prime ends are blocked to prevent unwanted priming sequencing begins with the extension of the first sequencing primer to produce the first read with each cycle for fluorescently tagged nucleotides compete for addition to the growing chain only one is incorporated based on the sequence of the template after
the addition of each nucleotide the clusters are excited by a light source and a characteristic fluorescent signal is emitted this proprietary process is called sequencing by synthesis the number of cycles determines the length of the read the emission wavelength along with the signal intensity determine the base call for a given cluster all identical strands are read simultaneously hundreds of millions of clusters are sequenced in a massively parallel process this image represents a small fraction of the flow cell after the completion of the first read the read product is washed away in this step the index
one read primer is introduced and hybridized to the template the read is generated similar to the first read after completion of the index read the read product is washed off and the three prime end of the template is d protected the template now folds over and binds the second ala go on the flow cell index to is read in the same manner as index one index to read product is washed off at the completion of this step polymerase Asst extend the second flow cell all ago forming a double-stranded bridge this double-stranded DNA is then linearized
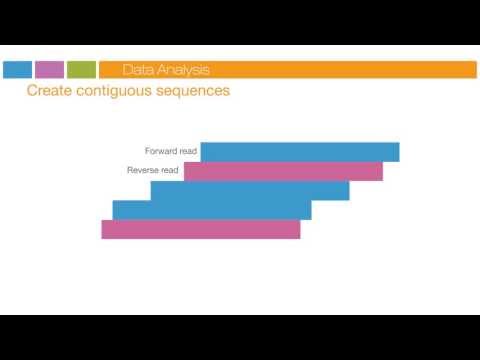
and the three prime ends blocked the original forward strand is cleaved off and washed away leaving the reverse strand read to begins with the introduction of the read two sequencing primer as with read one the sequencing steps are repeated until the desired read length is achieved the read to product is washed away this entire process generates billions of reads representing all the fragments sequences from pooled sample libraries are separated based on the unique indices introduced during the sample preparation for each sample reads with similar stretches of base calls are locally clustered forward and reverse reads
are paired creating contiguous sequences these contiguous sequences are aligned back to the reference genome for variant identification the paired end information is used to resolve ambiguous alignments you
Related Videos

5:13
Overview of Illumina Sequencing by Synthes...
Illumina
2,228,223 views

7:38
Next Generation Sequencing - A Step-By-Ste...
ClevaLab
307,958 views

4:09
Illumina sequencing – How it works
INTEGRA Biosciences
10,751 views

18:26
StatQuest: A gentle introduction to RNA-seq
StatQuest with Josh Starmer
493,856 views
4:23
Intro to Sequencing by Synthesis: Industry...
Illumina
331,845 views
4:44
Next Generation Sequencing (Illumina) - An...
Henrik's Lab
582,436 views

51:48
Illumina Sequencing Overview: Library Prep...
Ambry Genetics
176,514 views
41:35
Learn about Illumina's Next-Generation Seq...
Illumina
76,246 views
17:41
Single cell RNA sequencing overview | ScRN...
Animated biology With arpan
14,324 views

10:42
Illumina sequencing | DNA sequencing by sy...
Shomu's Biology
269,309 views

12:04
Illumina MiSeq System: How to Start a Run
Illumina
25,658 views

3:24
Ion Torrent Next-generation Sequencing
Thermo Fisher Scientific
175,669 views

23:44
Elon Musk's Bionic Eyes Are Here.
Michael Chua, MD
1,116,408 views

12:42
A Guide to Next Generation Sequencing Basi...
Bioinformagician
7,833 views

25:05
Next Generation Sequencing 2: Illumina NGS...
iBiology Techniques
158,558 views
43:32
Illumina | Introduction to Sequencing Data...
Illumina
24,513 views

31:26
Next Generation Sequencing 1: Overview - E...
iBiology Techniques
460,773 views

16:04
Next Gen SOLiD DNA Sequencing Method Expla...
"Special" Operations
110,715 views

7:03
4) Next Generation Sequencing (NGS) - Data...
Applied Biological Materials - abm
153,002 views

2:02
Illumina | Patterned Flow Cell Technology
Illumina
118,282 views