CRISPR-Cas9 Genome Editing Technology
643.29k views1840 WordsCopy TextShare
Professor Dave Explains
We've learned about a few techniques in biotechnology already, but the CRISPR-Cas9 system is one of ...
Video Transcript:
We’ve examined a handful of biotechnology concepts in previous tutorials, but now it’s time to introduce what is undoubtedly the most promising technique in biotechnology of the past decade. The CRISPR-Cas9 system represents genome editing technology that has revolutionized molecular biology, due to its precise and site-specific gene editing capabilities, which essentially allow for an unprecedented level of control in manipulating the genetic information of a living organism. How does this work mechanistically, and what are its applications?
Let’s get a closer look now, starting with some historical context. In 1987, Atsuo Nakata and his team of researchers from the Osaka University in Japan first reported the presence of Clustered Regularly Interspaced Short Palindromic Repeats, abbreviated as CRISPR, in the Escherichia coli genome. These refer to short, repeated sequences of DNA nucleotides found within the genome of prokaryotes.
These sequences are the same when read from 5' to 3' on one strand of DNA and from 5' to 3' on the complementary strand, and are therefore described as palindromic repeats, just the way that we refer to words like racecar or kayak as being palindromes, because they are the same whether read forwards or backwards. This was further reported in both Gram-positive and Gram-negative bacteria, along with archaea, leading to the obvious question regarding the relevance of CRISPR to these organisms, which drove research for some time. Later on, in the mid 2000s, the functionality and importance of CRISPR was first realized in prokaryotes.
As it turns out, the CRISPR system is a key component of their adaptive immunity, which protects these prokaryotes from attack by viral DNA, bacteriophages, and plasmids. That’s right, it may seem incredible, but even unicellular bacteria have a very basic immune system. Recall from our studies in the immunology series that adaptive immunity refers to the immunity that an organism acquires after exposure to an antigen, either from a pathogen or vaccination.
Vaccination, for example, results in a form of adaptive immunity in humans, since the body is exposed to antigens, and forms antibodies in response, which contribute to the development of the immunity. The way this works for bacteria is as follows. The unique sequences that are nestled in between the palindromic repeats, which are called spacers, are bits of DNA that are foreign, and do not belong to the bacterium, but instead originate from mobile genetic elements, or MGEs, such as bacteriophages, transposons, or plasmids that have previously infected the prokaryote.
This was revealed by sequencing the spacers found in the CRISPR system, which led to the hypothesis that this could be a defense mechanism employed by bacteria to recognize foreign DNA elements. During a viral infection, bacteria acquire a small piece of the foreign viral DNA, and integrate it into the CRISPR locus to generate CRISPR arrays. These consist of duplicate sequences, which are the palindromic repeats belonging to the bacterial genome, flanked by variable sequences, or spacers, which again are from the foreign genetic elements.
In this way, bacteria retain a memory, so to speak, of a past infection. So although it was initially revealed as a genomic component of bacteria and archaea, CRISPR has inspired a method of genome editing that can be applied to various eukaryotic species. But before we get there, we first have to understand the function of CRISPR in prokaryotes, because understanding the mechanism of its natural function will be necessary in order to understand the way it is exploited to achieve genome editing capabilities in humans and other organisms.
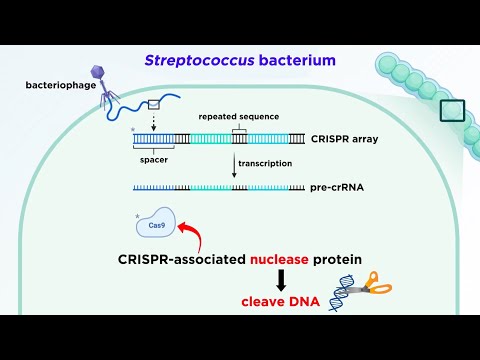
Let’s take a look at a particular Streptococcus bacterium which is being attacked by a bacteriophage. Once the viral DNA is injected into the cell, a section of it can be incorporated into the bacterial genome, and as we mentioned, it will be inserted between the repeated palindromic sequences. This will now be called a spacer.
So here we can see three different spacers, potentially from three different viruses, sandwiched in between the repeated palindromic sequences. Now we have what is called a CRISPR array. This CRISPR array can undergo transcription, to form CRISPR RNA, abbreviated as crRNA, although this longer strand is called pre-crRNA.
Then the protein Cas9 gets involved. Cas refers to CRISPR-associated nuclease protein, and as we know, nucleases are enzymes that are capable of cleaving DNA at specific nucleotide linkages, kind of like a pair of scissors. In particular, Cas9 is one of the nucleases found in Streptococcus pyogenes, which is one of the most extensively researched and characterized CRISPR-associated nuclease proteins, so this is the one we will be looking at here inside this bacterium.
Now along with Cas9, there are also molecules of tracrRNA. These have sections that are complementary to and therefore can anneal to the palindromic repeats. So for each spacer and palindromic repeat, we end up with a complex consisting of that segment of pre-crRNA, a tracrRNA, and a Cas9 protein.
Then another enzyme called ribonuclease three, or RNase III, will cleave the strand in between these complexes, leaving us with individual crRNA complexes which we can call effector complexes. With these effector complexes formed, the cell is now ready to defend against the invader whose genome produced that crRNA. If this complex encounters a section of viral DNA that has a sequence which is complementary to this crRNA, the nuclease enzyme will coordinate, and if it recognizes a short sequence unique to the viral genome called a protospacer adjacent motif, or PAM, then it will snip both strands of the DNA, just a few base pairs upstream from the PAM.
In doing so, it will neutralize the virus, because its genome can no longer be transcribed properly to create more viral particles, so infection is impossible. So that gives us a reasonable understanding of how CRISPR is employed by prokaryotic organisms as a natural defense. Now it’s time to understand how this phenomenon came to serve as the basis for biotechnological application.
This begins in 2012, when Jennifer Doudna, a molecular biologist from the University of California, Berkeley along with French microbiologist Emmanuelle Charpentier, were the first to propose that the bacterial CRISPR-Cas9 system could be used as a programmable toolkit for genome editing in humans and other animal species, and they eventually received the Nobel prize in chemistry for their work, in 2020. So how can genome editing be achieved using this method? The first thing we need to understand is that in bacteria, the crRNA and tracrRNA are separate molecular entities.
The first major breakthrough arrived when it was realized that the roles of these molecules could be combined into a single molecule by fusing them together with a linker to generate something called single guide RNA, or sgRNA, which can be synthesized in the lab. If the sgRNA complexes with a Cas9 protein, this two-component system will be able to cleave DNA just as the three-component system does in bacteria. What this means was that it was then possible to determine any sequence of about 20 base pairs as a target for editing, and all that has to be done is to synthesize the appropriate sgRNA with the complementary sequence, and insert that into a cell along with the Cas9 protein which has been sourced from Streptococcus pyogenes.
The complex will form, read the DNA until it finds the appropriate sequence along with a PAM sequence, binding will occur, and DNA will be cleaved at precisely the desired location. Cas9 has two domains, and each one will snip one of the DNA strands. After the incision is made, the natural DNA repair mechanism is enacted for the target DNA.
The cleaved dsDNA can undergo repair via two routes. Either by homology-directed repair, abbreviated as HDR, or by non-homologous end joining, abbreviated as NHEJ. The NHEJ pathway repairs double-strand breaks in DNA by directly ligating without the need for a homologous template, which means a DNA strand with similar sequence that can act as a template.
The NHEJ mechanism can also introduce insertion or deletion of specific sequences at the joining ends, thus creating what are referred to as indels. Indels are DNA strands with either an insertion or deletion of nucleotide sequences. Thus, NHEJ produces DNA strands with non-uniformity in size.
The other route of repair, the HDR pathway, is commonly found in bacterial and archaeal cells, while the NHEJ pathway we just discussed is more common in a eukaryotic domain. The HDR process, although more complex than NHEJ, uses a homologous DNA template. The homologous DNA template has homology to the adjacent sequences surrounding the site of cleavage to incorporate new DNA fragments.
The template guides the repair process, and lowers the possibility of errors. Since there is no insertion or deletion of nucleotide sequences, the HDR pathway maintains uniformity in the size of the resulting dsDNA, unlike NHEJ. So that covers the mechanism of CRISPR genome editing technology.
Now we move on to the potential applications, which have only expanded ever since Doudna and Charpentier suggested the possibility of using CRISPR for genome editing in humans and other animals. The potential scope of application of CRISPR is vast, and includes its use as a genetic screen to identify genes in different cells. One of the most prominent applications is in cancer immunotherapy.
In this practice, immune T cells, which are a type of white blood cell that fights against a disease, are genetically modified using CRISPR technology. Specifically, these T cells are extracted from the patient’s body and modified to make them more specialized in recognizing cancer cells and killing them once they are reintroduced into the patient’s body. Similarly, CRISPR has also found its application in therapeutic management of acquired immunodeficiency syndrome, or AIDS, which is caused by human immunodeficiency virus, also known as HIV, as we covered in the microbiology series.
Conventional anti-retroviral therapies are capable of suppressing viral replication. But once the virus gets converted to its proviral form, conventional therapies are ineffective in targeting the virus. The provirus resides within the immune cells and continues to make copies of itself using the immune cell machinery, and the immune cells fail to target the proviral latent reservoir which presents the risk of viral rebound or relapse of the disease.
Other than cancer and AIDS, CRISPR has also found immense application in developing assays to detect SARS-CoV-2 infection, the cause of the current global pandemic. Although genome editing of human embryos and their implantation into a human womb, as well as genetic editing of somatic cells, have wide ethical concerns and potential risks, CRISPR has the promise to cure various diseases and prevent the inheritance of gene-linked diseases. Additionally, genome editing in plants using CRISPR technology introduces the possibility of making plants resistant to certain diseases, improving their phenotype or observable characteristics, incorporating certain specific traits, improving crop yield, and so forth.
With so many invigorating possibilities for this exciting new technology, it will be fascinating to see which of these major diseases and issues will be solved first, signaling the dawn of a new era in molecular biology.
Related Videos

35:15
Understanding CRISPR-Cas9
Andrew Douch
173,935 views

10:02
But what is CRISPR-Cas9? An animated intro...
Powerhouse of the Cell
328,973 views

12:59
The First CRISPR Gene Therapy Is Here
SciShow
765,236 views
7:21
What is CRISPR?
Bozeman Science
1,925,334 views
5:29
How CRISPR lets you edit DNA - Andrea M. H...
TED-Ed
1,440,665 views

7:37
CRISPR's Next Advance Is Bigger Than You T...
TED
787,946 views

34:30
The Definitive Guide to Debunking Creation...
Professor Dave Explains
161,906 views

13:00
Gene Silencing Methods: CRISPR vs. TALENs ...
Applied Biological Materials - abm
175,347 views

15:54
How CRISPR lets us edit our DNA | Jennifer...
TED
1,814,351 views

36:06
The Definitive Guide to Debunking Creation...
Professor Dave Explains
50,951 views

1:22:46
Neanderthal Genome Project: Insights into ...
Linda Hall Library
169,226 views

2:01:21
GENE EDITING EXPLAINED! - A comprehensive ...
Genomics Guru
19,838 views

48:26
Jennifer Doudna: CRISPR Basics
Innovative Genomics Institute – IGI
362,275 views
14:13
You've Been Lied To About Genetics
SubAnima
922,087 views

27:54
Designing gRNA Oligos to Clone into Cas9 E...
Jacob Elmer
59,039 views

16:53
Biologist Explains One Concept in 5 Levels...
WIRED
4,174,174 views

35:56
The Insane Biology of: The Tardigrade
Real Science
1,007,942 views

16:48
2️⃣ CRISPR-Cas9: come funziona il sistema ...
DNA a colazione
18,459 views

52:00
16. Recombinant DNA, Cloning, & Editing
MIT OpenCourseWare
68,371 views

1:26:30
Rewiring the Brain: The Promise and Peril ...
World Science Festival
677,549 views